library(brms)
library(ggplot2)
library(marginaleffects)
library(modelsummary)
options(brms.backend = "cmdstanr")
theme_set(theme_minimal())
titanic <- read.csv("https://vincentarelbundock.github.io/Rdatasets/csv/Stat2Data/Titanic.csv")
titanic <- subset(titanic, PClass != "*")
f <- Survived ~ SexCode + Age + PClass
mod_prior <- brm(f,
data = titanic,
prior = c(prior(normal(0, .2), class = b)),
cores = 4,
sample_prior = "only")
mod_posterior <- brm(f,
data = titanic,
cores = 4,
prior = c(prior(normal(0, .2), class = b)))Bayesians often advocate for the use of prior predictive checks (Gelman et al. 2020). The idea is to simulate from the model, without using the data, in order to refine the model before fitting. For example, we could draw parameter values from the priors, and use the model to simulate values of the outcome. Then, could inspect those to determine if the simulated outcomes (and thus the priors) make sense substantively. Prior predictive checks allow us to iterate on the model without looking at the data multiple times.
One major challenge lies in interpretation: When the parameters of a model are hard to interpret, the analyst will often need to transform before they can assess if the generated quantities make sense, and if the priors are an appropriate representation of available information.
In this post I show how to use the marginaleffects and brms packages for R to facilitate this process. The benefit of the approach described below is that it allows us to conduct prior predictive checks on the actual quantities of interest. For example, if the ultimate quantity that we want to estimate is a contrast or an Average Treatment Effect, then we can use marginaleffects to simulate the specific quantity of interest using just the priors and the model.
In this example, we create two model objects with brms. In one of them, we set sample_prior="only" to indicate that we do not want to use the dataset at all, and that we only want to use the priors and model for simulation:
Now, we use the avg_comparisons() function from the marginaleffects package to compute contrasts of interest:
cmp <- list(
"Prior" = avg_comparisons(mod_prior),
"Posterior" = avg_comparisons(mod_posterior))Finally, we compare the results with and without the data in tables and plots:
modelsummary(
cmp,
output = "markdown",
statistic = "conf.int",
fmt = fmt_significant(2),
gof_map = NA,
shape = term : contrast ~ model)| Prior | Posterior | |
|---|---|---|
| Age +1 | 0.005 | -0.0058 |
| [-0.391, 0.396] | [-0.0080, -0.0037] | |
| PClass 2nd - 1st | -0.00099 | -0.19 |
| [-0.39570, 0.39466] | [-0.27, -0.12] | |
| PClass 3rd - 1st | 0.0036 | -0.38 |
| [-0.3841, 0.3907] | [-0.45, -0.31] | |
| SexCode 1 - 0 | 0.0072 | 0.49 |
| [-0.3933, 0.3916] | [0.44, 0.55] |
draws <- lapply(names(cmp), \(x) transform(get_draws(cmp[[x]]), Label = x))
draws <- do.call("rbind", draws)
ggplot(draws, aes(x = draw, color = Label)) +
xlim(c(-1, 1)) +
geom_density() +
facet_wrap(~term + contrast, scales = "free")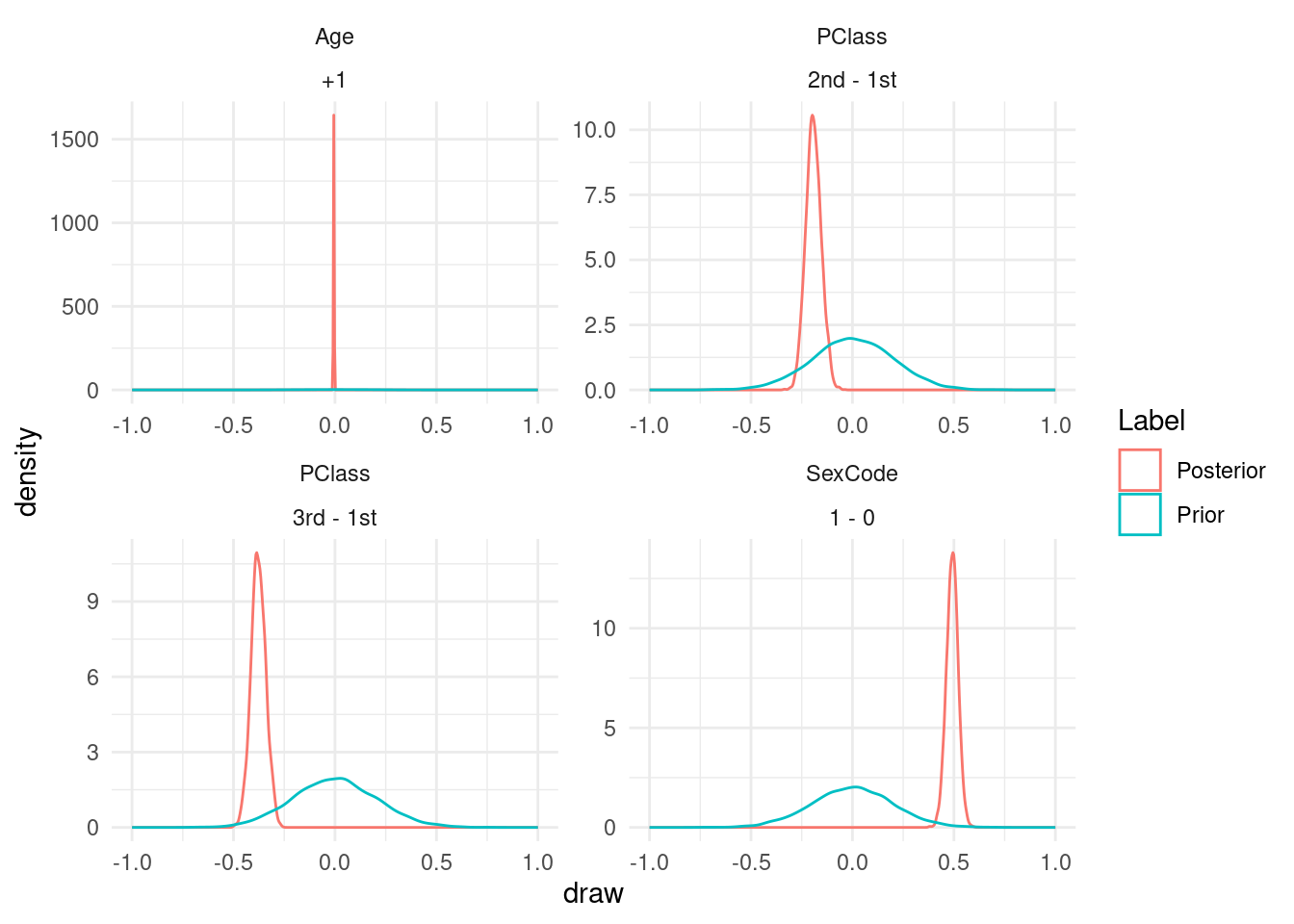
This kind of approach is particularly useful with more complicated models, such as this one with categorical outcomes. In such models, it would be hard to know if a normal prior is appropriate for the different parameters:
modcat_posterior <- brm(
PClass ~ SexCode + Age,
prior = c(
prior(normal(0, 3), class = b, dpar = "mu2nd"),
prior(normal(0, 3), class = b, dpar = "mu3rd")),
family = categorical(link = logit),
cores = 4,
data = titanic)
modcat_prior <- brm(
PClass ~ SexCode + Age,
prior = c(
prior(normal(0, 3), class = b, dpar = "mu2nd"),
prior(normal(0, 3), class = b, dpar = "mu3rd")),
family = categorical(link = logit),
sample_prior = "only",
cores = 4,
data = titanic)pd <- get_draws(comparisons(modcat_prior))
comparisons(modcat_prior) |> summary() rowid term group contrast
Min. : 1.0 Length:4536 Length:4536 Length:4536
1st Qu.:189.8 Class :character Class :character Class :character
Median :378.5 Mode :character Mode :character Mode :character
Mean :378.5
3rd Qu.:567.2
Max. :756.0
estimate conf.low conf.high
Min. :-3.546e-03 Min. :-0.8241741 Min. :0.0000188
1st Qu.: 0.000e+00 1st Qu.:-0.3322749 1st Qu.:0.0162391
Median : 0.000e+00 Median :-0.0947103 Median :0.0917383
Mean : 4.690e-06 Mean :-0.2005252 Mean :0.2005384
3rd Qu.: 0.000e+00 3rd Qu.:-0.0157481 3rd Qu.:0.3328813
Max. : 4.111e-03 Max. :-0.0000197 Max. :0.8134400
predicted_lo predicted_hi predicted tmp_idx
Min. :0.000e+00 Min. :0.000e+00 Min. :0.000e+00 Min. : 1.0
1st Qu.:0.000e+00 1st Qu.:0.000e+00 1st Qu.:0.000e+00 1st Qu.:189.8
Median :1.017e-05 Median :1.466e-05 Median :1.017e-05 Median :378.5
Mean :1.592e-02 Mean :1.571e-02 Mean :1.574e-02 Mean :378.5
3rd Qu.:2.471e-03 3rd Qu.:2.550e-03 3rd Qu.:2.471e-03 3rd Qu.:567.2
Max. :1.966e-01 Max. :1.966e-01 Max. :1.966e-01 Max. :756.0
PClass SexCode Age
Length:4536 Min. :0.000 Min. : 0.17
Class :character 1st Qu.:0.000 1st Qu.:21.00
Mode :character Median :0.000 Median :28.00
Mean :0.381 Mean :30.40
3rd Qu.:1.000 3rd Qu.:39.00
Max. :1.000 Max. :71.00 comparisons(modcat_posterior) |> summary() rowid term group contrast
Min. : 1.0 Length:4536 Length:4536 Length:4536
1st Qu.:189.8 Class :character Class :character Class :character
Median :378.5 Mode :character Mode :character Mode :character
Mean :378.5
3rd Qu.:567.2
Max. :756.0
estimate conf.low conf.high predicted_lo
Min. :-1.407e-01 Min. :-0.218159 Min. :-0.065904 Min. :0.02497
1st Qu.:-1.223e-02 1st Qu.:-0.039564 1st Qu.:-0.008224 1st Qu.:0.21641
Median : 5.173e-04 Median :-0.008402 Median : 0.005425 Median :0.29599
Mean : 7.790e-06 Mean :-0.034669 Mean : 0.034797 Mean :0.33287
3rd Qu.: 2.658e-02 3rd Qu.: 0.010558 3rd Qu.: 0.090094 3rd Qu.:0.45914
Max. : 1.401e-01 Max. : 0.051580 Max. : 0.225256 Max. :0.89558
predicted_hi predicted tmp_idx PClass
Min. :0.02701 Min. :0.02529 Min. : 1.0 Length:4536
1st Qu.:0.22373 1st Qu.:0.21641 1st Qu.:189.8 Class :character
Median :0.31450 Median :0.29850 Median :378.5 Mode :character
Mean :0.33275 Mean :0.33283 Mean :378.5
3rd Qu.:0.42014 3rd Qu.:0.44022 3rd Qu.:567.2
Max. :0.90812 Max. :0.89558 Max. :756.0
SexCode Age
Min. :0.000 Min. : 0.17
1st Qu.:0.000 1st Qu.:21.00
Median :0.000 Median :28.00
Mean :0.381 Mean :30.40
3rd Qu.:1.000 3rd Qu.:39.00
Max. :1.000 Max. :71.00