library(data.table)
library(fixest)This is another post in my series of attempts to learn the data.table package for R and to get more comfortable with base R graphics. Today, I reimplement the data generating process and one of the models in this excellent blog post by Andrew Baker: How to create relative time indicators. I also strongly recommend watching Andrew’s talk on Difference-in-Differences staggered treatment designs.
For this exercise, we will only use two libraries:
As in the original post, the data generating process includes unit and time fixed effects, as well as unit-specific treatment effects.
\[y_{it}=\alpha_i + \alpha_t + \tau_{it} + \varepsilon_{it}\] \[\alpha_i,\alpha_t \sim N(0,1)\] \[\varepsilon_{it} \sim N(0,0.5)\] \[\mu_{it} \sim N(0.3,0.2^2)\]
and \(\tau_{it}\) is equal to the sum of each firm-specific \(\mu_{it}\) since the start of the treatment period.
We have 1000 firms split into 4 cohorts which receive treatment in 1986, 1992, 1998, and 2004.
make_data = function() {
# unit fixed effects
unit = data.table(
unit = 1:1000,
unit_fe = rnorm(1000, 0, 1), # unit fe
state = sample(1:40, 1000, replace=TRUE), # state id
mu = rnorm(1000, .3, .2) # treatment effect
)
# year fixed effects
year = data.table(
year = 1980:2010,
year_fe = rnorm(31, 0, 1)
)
# treatment groups by state
treat_taus = data.table(
state = sample(1:40, 40),
cohort_year = sort(rep(c(1986, 1992, 1998, 2004), 10))
)
# full interaction of unit X year
out = CJ(
unit = 1:1000,
year = 1980:2010
)
out = merge(out, unit, by="unit")
out = merge(out, year, by="year")
out = merge(out, treat_taus, by="state")
# error term, treatment indicator, and treatment effects
out[, error := rnorm(31000, 0, .5)][
, treat := fifelse(year >= cohort_year, 1, 0)][
, tau := fifelse(treat == 1, mu, 0)]
# cumulative treatment effects
setkey(out, unit, year)
out[, tau_cum := cumsum(tau), by="unit"]
# dependent variable
out[, dep_var := unit_fe + year_fe + tau_cum + error]
return(out)
}Plot data from a single simulation
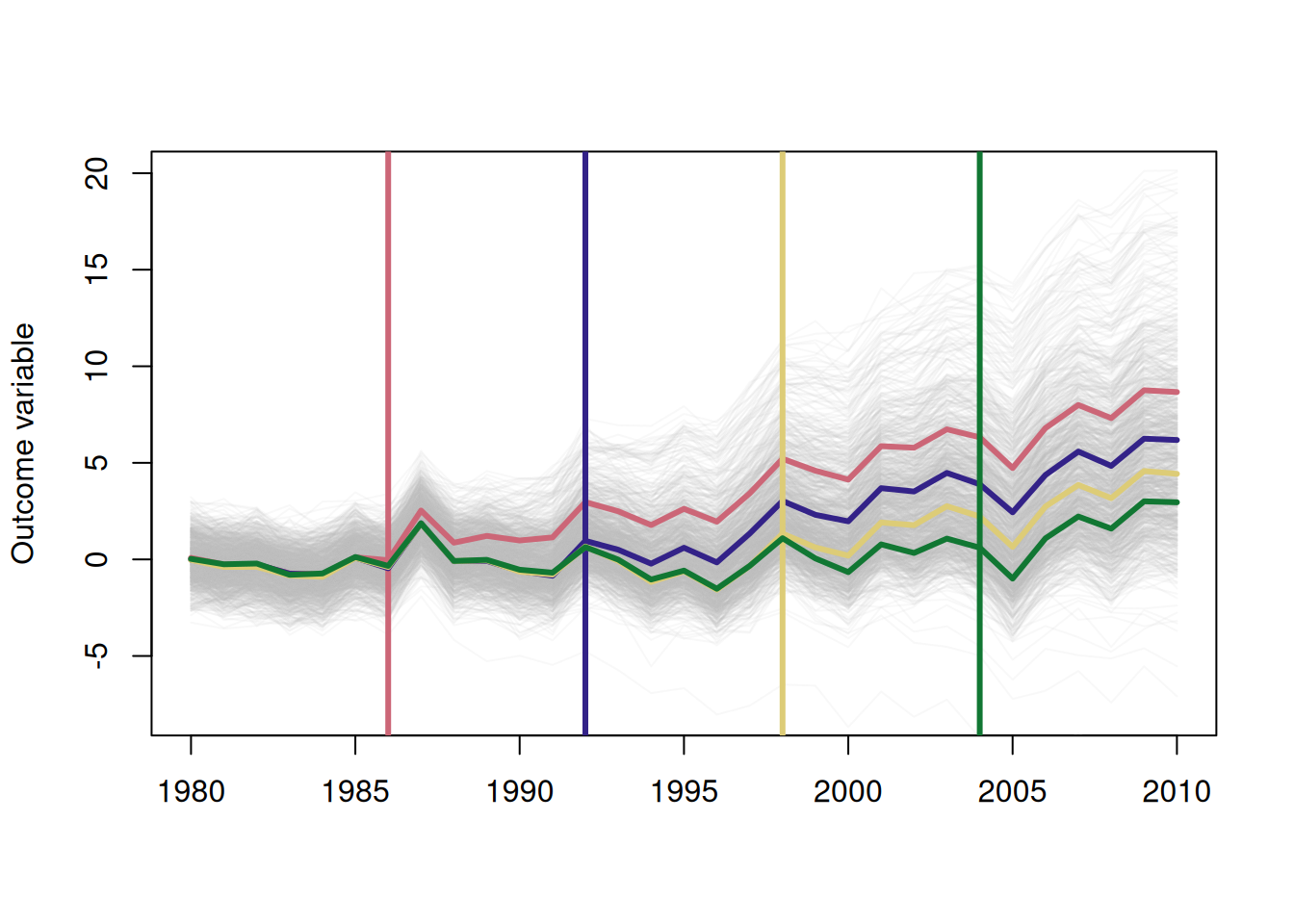
dat = make_data()
# empty plot window
plot(0,
type = "n",
ylim = c(-8, 20),
xlim = c(1980, 2010),
xlab = "",
ylab = "Outcome variable")
# one line per firm
for (i in 1:1000) {
lines(x = dat[unit==i]$year,
y = dat[unit==i]$dep_var,
col = adjustcolor("grey", alpha=.1))
}
# means by treatment cohort
cohorts = dat[, .(dep_var = mean(dep_var)), by=.(cohort_year, year)]
years <- c(
"#CC6677" = 1986,
"#332288" = 1992,
"#DDCC77" = 1998,
"#117733" = 2004
)
for (y in years) {
abline(v = y,
col = names(years)[years==y],
lwd = 3)
lines(x = cohorts[cohort_year==y]$year,
y = cohorts[cohort_year==y]$dep_var,
col = names(years)[years==y],
lwd = 3)
}
# simulate data
dat = make_data()
# drop 2004 treatment
dat = dat[year <= 2003]
# years since/to treatment
dat[, rel_year := year - cohort_year]
min_year = dat[cohort_year==2004, min(rel_year)]
max_year = dat[cohort_year==2004, max(rel_year)]
# identification requires dropping two dummies
# dat[, rel_year := fifelse(!rel_year %in% c(min_year, -1), as.character(rel_year), "omitted")][
# , rel_year := factor(rel_year)][
# , rel_year := relevel(rel_year, ref=c("omitted")]
dat[, rel_year := factor(rel_year, sort(unique(rel_year)))]
# regression model
f = dep_var ~ rel_year | unit + year
mod = feols(f, data=dat)The variables 'rel_year12', 'rel_year13', 'rel_year14', 'rel_year15', 'rel_year16' and 'rel_year17' have been removed because of collinearity (see $collin.var).# clean results
out = data.table("term" = names(coef(mod)), "estimate" = coef(mod))
out = out[ , term := as.numeric(gsub(".*year", "", term))]
out = out[term %in% -5:5]Plot results
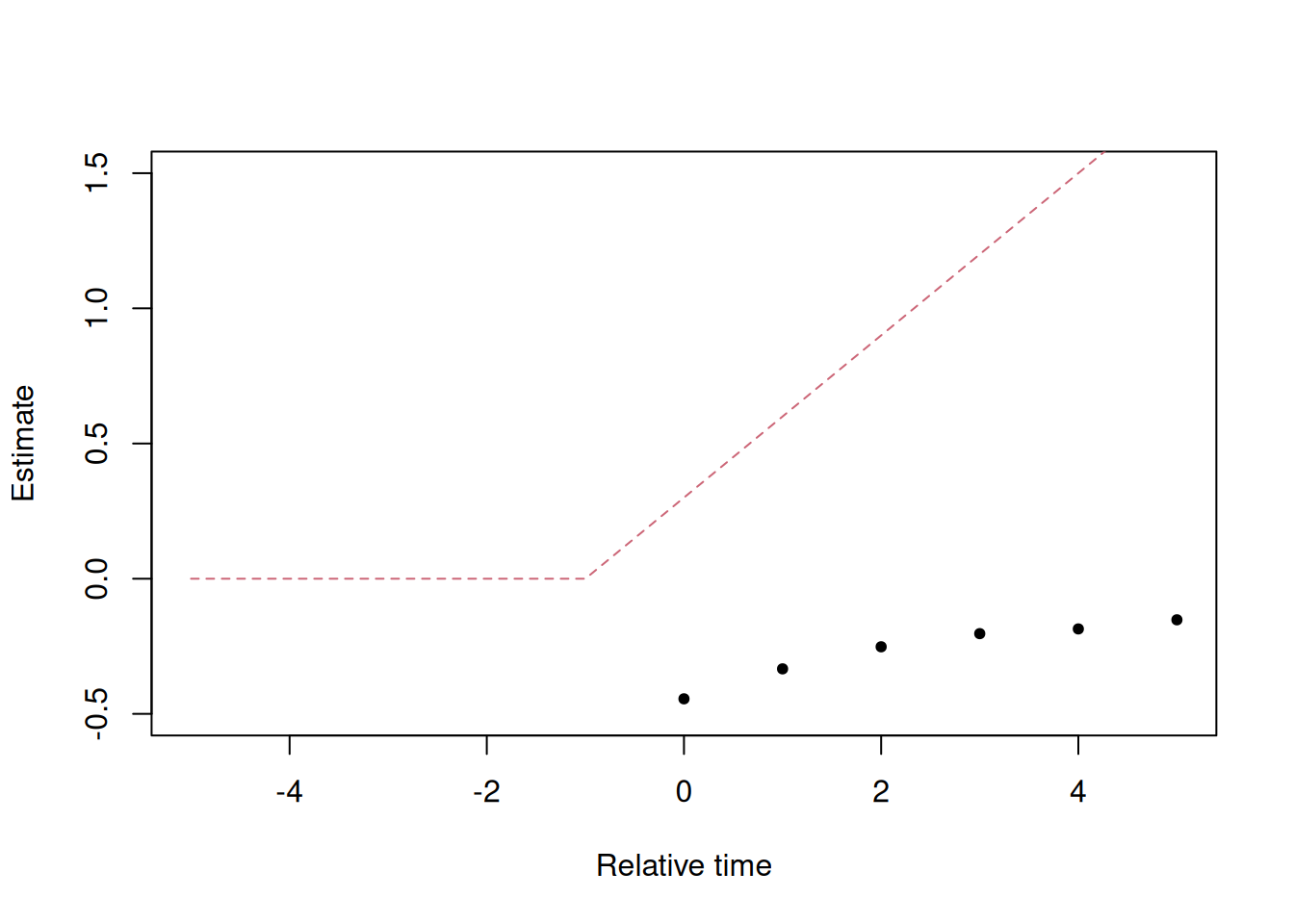
# event study estimates
plot(x = out$term,
y = out$estimate,
pch=20,
xlim = c(-5, 5),
ylim = c(-.5, 1.5),
xlab = "Relative time",
ylab = "Estimate")
# truth
truth = data.table(term=-5:5)
truth[, estimate := fifelse(term >= 0, (term + 1) * .3, 0)]
lines(x = truth$term,
y = truth$estimate,
col = "#CC6677",
lty = "dashed")