f <- function(x, y) {
if (x > y) {
return(x)
} else {
return(y)
}
}This notebook illustrates how one can apply a function to every row of a dataset using the data.table package. I begin with a trivial example, and continue with a slightly less trivial one.
Trivial example
Say you want to apply this function:
to every row of this dataset:
library(data.table)
set.seed(6913)
dat <- data.table(x = sample(1:10, 4),
y = sample(1:10, 4))
dat x y
<int> <int>
1: 9 1
2: 2 4
3: 3 9
4: 1 6Since f() is not vectorized, you can’t simply pass the x and y vectors to f(). This would produce a warning and the wrong answer:
#dat[, z := f(x, y)][]Instead, we will explore 2 alternatives for rowwise operations in data.table, using the by argument or Map function. In the end, our code will look like this:
dat[, z1 := f(x, y), by=1:nrow(dat)]
dat[, z2 := Map(f, x, y)]
dat x y z1 z2
<int> <int> <int> <list>
1: 9 1 9 9
2: 2 4 4 4
3: 3 9 9 9
4: 1 6 6 6In the code above, by=1:nrow(dat) tells data.table that it should treat every row as a distinct group, and that it should apply the f function to each of those groups separately. The Map function (from base R) applies an element-wise operation to the “x” and “y” vectors, and returns a list.
To understand how these two strategies work, we will now consider a slightly more complicated (but still rather trivial) example.
Task description
Consider this data generating process:
\[y = 1 \cdot x + \varepsilon\] \[z = \beta_{xz}\cdot x + \beta_{yz}\cdot y + \nu,\]
where \(\varepsilon, \nu, x \sim \mbox{Normal}(0, 1)\). To assess the effect of \(x\) on \(y\), we can estimate a linear regression model with \(y\) as dependent variable and \(x\) as regressor. In R, we could type lm(y~x). However, if we estimate a more complex model with z as control variable, lm(y ~ x + z), our estimates are likely to be biased, because z is “post-treatment” with respect to x. In DAG-world, they call this “collider bias”.
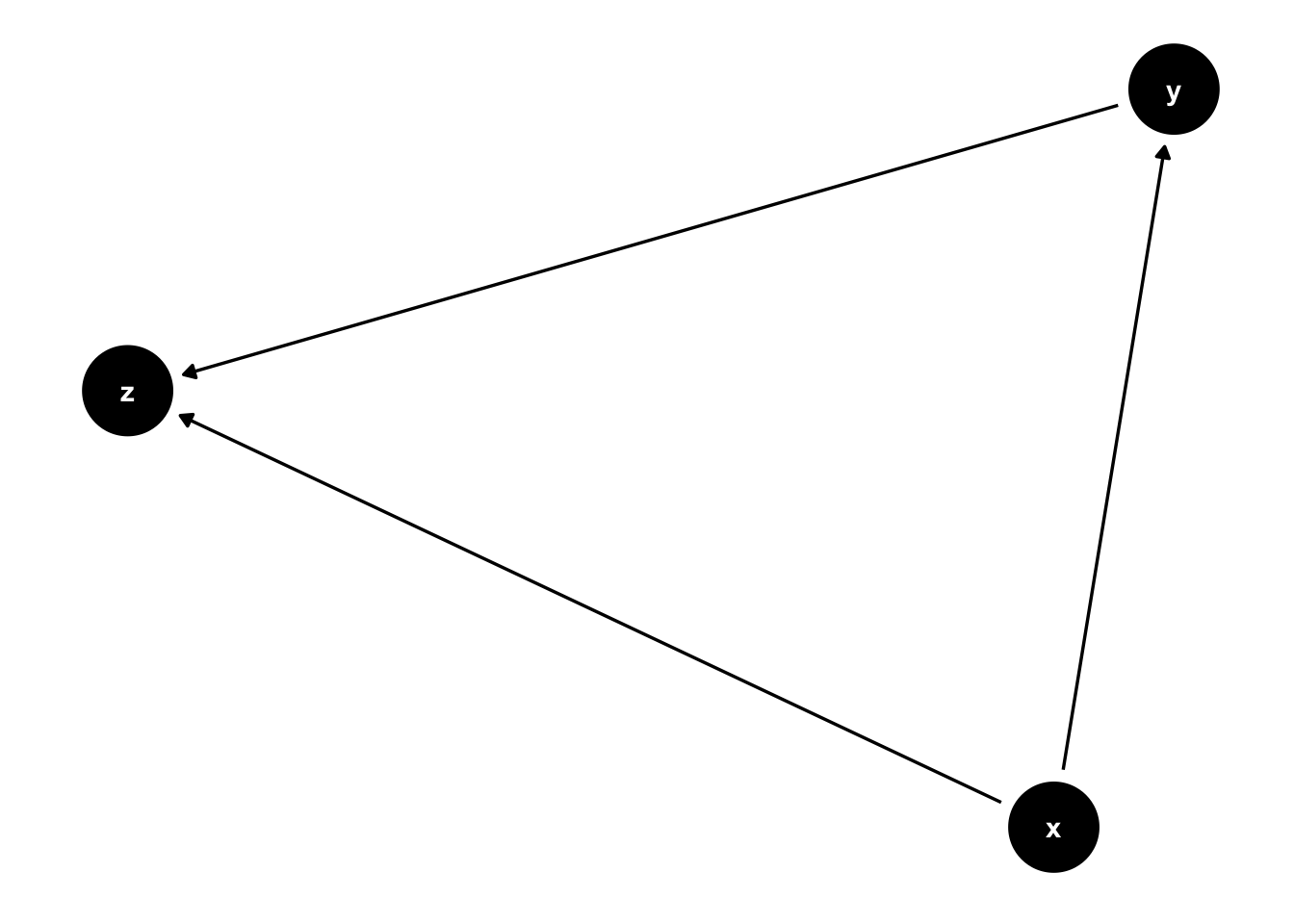
Our task will be to estimate the magnitude of this collider bias for different values of \(\beta_{yz}\) and \(\beta_{xz}\) using rowwise operations in data.table.
A partial solution
First, we define a function to simulate datasets which conform to the data generating process described above:
simulation <- function(b_zx, b_zy) {
N <- 100 # 100 observations
x <- rnorm(N) # x has no ancestors
y <- 1 * x + rnorm(N) # x causes y
z <- b_zx * x + b_zy * y + rnorm(N) # x and y cause z
out <- list("Biased"=coef(lm(y~x+z))["x"], # collider bias
"Unbiased"=coef(lm(y~x))["x"]) # unbiased model
out
}Note that this function returns two “x” coefficients in a list. This is important because data.table treats elements of lists as distinct columns. This ensures that our final output will a dataset with a column called “Biased” and a column called “Unbiased”.
We want to assess the performance of both models for every combination of b_xz and b_yz in {-1, 0, 1}, and we want to replicate our experiment 100 times. To achieve this, we use the CJ function to create a dataset with every combination of b_xz, b_yz, and a replication index:
params <- CJ(replication = 1:100,
b_xz = -1:1,
b_yz = -1:1)
dim(params)[1] 900 3tail(params)Key: <replication, b_xz, b_yz>
replication b_xz b_yz
<int> <int> <int>
1: 100 0 -1
2: 100 0 0
3: 100 0 1
4: 100 1 -1
5: 100 1 0
6: 100 1 1The simulation function is not vectorized, which means that it only accepts 2 numerical values as arguments; we can’t simply give it the full vectors of parameters in params. We need to apply the simulation function rowwise, that is, to every row of the dataset. To achieve this, we use data.table’s by argument. Typically, by is used to apply an operation to a “group” or “subset” of the data. Here, we assign each row to its own group:
results <- params[, simulation(b_xz, b_yz), by=1:nrow(params)]
head(results) nrow Biased Unbiased
<int> <num> <num>
1: 1 -0.01723128 0.9884276
2: 2 0.92631128 1.0146390
3: 3 1.03765059 1.0041715
4: 4 0.28818854 0.9350003
5: 5 0.91518233 0.9129573
6: 6 0.61884030 1.2013585Great! Now we know that the unbiased estimate for row 1 is equal to -0.0172313, and that the unbiased estimate for row 1 is equal to 0.9884276. The problem, of course, is that “row 1” is not very informative. We lost relevant information about the values of b_yz and b_xz on every row.
A better solution
A better way to leverage the by argument would be:
results <- params[, simulation(b_xz, b_yz), by=list(replication, b_xz, b_yz)]
head(results)Key: <replication, b_xz, b_yz>
replication b_xz b_yz Biased Unbiased
<int> <int> <int> <num> <num>
1: 1 -1 -1 0.01247715 1.0371168
2: 1 -1 0 1.03431370 0.9687347
3: 1 -1 1 1.07899244 0.9738551
4: 1 0 -1 0.72669444 1.1135464
5: 1 0 0 0.97200903 0.9751390
6: 1 0 1 0.70268068 1.1879717Now we can find the mean biased and unbiased estimates using normal data.table operations:
results[, list(Biased=mean(Biased), Unbiased=mean(Unbiased)), by=list(b_xz, b_yz)][] b_xz b_yz Biased Unbiased
<int> <int> <num> <num>
1: -1 -1 0.005753066 1.0060821
2: -1 0 1.032445223 1.0195756
3: -1 1 0.998845382 0.9945345
4: 0 -1 0.510010117 0.9965415
5: 0 0 0.990174328 0.9907054
6: 0 1 0.497241640 0.9976653
7: 1 -1 1.006002068 1.0077494
8: 1 0 1.003550065 0.9986808
9: 1 1 0.004560044 0.9867779Recall that the true value of the coefficient is 1. There appears to be no collider bias (a) x does not cause z, (b) y does not cause z, or (b) the effects of x and y on z exactly offset each other (-1 and 1).
(These are weird edge cases. Don’t condition on colliders!)
A useful alternative
An alternative approach is to use the Map function from base R. The Map(simulate, x, y) call produces a list where element 1 is simulate(x[1], y[1]), element 2 is simulate(x[2], y[2]), etc. This list can be used as a list-column in a “nesting” / “unnesting” programming pattern:
params[, coefs := Map(simulation, b_xz, b_yz)]
head(params)Key: <replication, b_xz, b_yz>
replication b_xz b_yz coefs
<int> <int> <int> <list>
1: 1 -1 -1 <list[2]>
2: 1 -1 0 <list[2]>
3: 1 -1 1 <list[2]>
4: 1 0 -1 <list[2]>
5: 1 0 0 <list[2]>
6: 1 0 1 <list[2]>Notice that each entry in the coefs column is itself a list of coefficients. We can “unnest” that list:
params[, coefs[[1]], by=list(replication, b_xz, b_yz)]Key: <replication, b_xz, b_yz>
replication b_xz b_yz Biased Unbiased
<int> <int> <int> <num> <num>
1: 1 -1 -1 -0.1548834 0.9964310
2: 1 -1 0 1.1573618 1.0512472
3: 1 -1 1 1.0874509 1.1651031
4: 1 0 -1 0.4060620 0.9381167
5: 1 0 0 1.0408373 1.0438923
---
896: 100 0 0 0.9937826 0.9890470
897: 100 0 1 0.5487397 1.1051524
898: 100 1 -1 0.9404295 1.0078137
899: 100 1 0 1.0258530 1.0156454
900: 100 1 1 -0.2287936 0.9910195Benchmarking
To optimize efficiency, the first thing to consider is whether you actually need to apply a function to every row. In general, this is a very expensive operation, and you are better off using a vectorized approach.
In most rowwise applications, the computational difference between the two approaches outlined above is likely to dwarfed by the cost of computation inside the function you apply rowwise. My advice is to just use the approach you think is easier to read and understand.
library(microbenchmark)
params <- CJ(replication = 1:100,
b_xz = -1:1,
b_yz = -1:1)
f_map <- function() {
out <- params[, coefs := Map(simulation, b_xz, b_yz)]
out <- out[, coefs[[1]], by=list(b_xz, b_yz)]
out
}
f_by <- function() {
out <- params[, simulation(b_xz, b_yz), by=list(replication, b_xz, b_yz)]
out
}
microbenchmark(f_map(), f_by(), times=3)Warning in microbenchmark(f_map(), f_by(), times = 3): less accurate nanosecond
times to avoid potential integer overflowsUnit: milliseconds
expr min lq mean median uq max neval cld
f_map() 327.1575 327.703 328.1253 328.2484 328.6092 328.9700 3 a
f_by() 322.2630 328.103 333.2066 333.9431 338.6785 343.4139 3 a